Example
Example 1: Now let us return to the problem we posed in Example 1 of Three-way Contingency Tables, namely to find the most parsimonious model that fits the data. Figure 1 summarizes the key results for all 18 models.

Figure 1 – Summary of all log-linear regression models
From Figure 1, we see that (CGT), (CG, GT, CT), (CG, GT), (CG, CT) and (T, CG) are the only models that fit the data, i.e. provide expected cell frequencies that are not significantly different from the observed cell frequencies. We now describe how to determine which of these models is the most parsimonious.
Observation
If model M1 has chi-square statistic (maximum likelihood) of χ12 with df1 degrees of freedom and model M2 has chi-square statistic (maximum likelihood) of χ22 with df2 degrees of freedom, then
Suppose M1 and M2 both provide a significant fit for the observed data and there is no significant difference between χ12 and χ22 then if M2 is a simpler model than M1 we could just as well use M2 to model our data as M1.
Example (continued)
Clearly the saturated model (CGT) is a perfect fit for our data, but, using the above observation, we would like to find a simpler model (one with fewer terms) that is not significantly different from the saturated model.
Of the five models that fit the data, the next most complex model is (CG, GT, CT). The difference between the chi-square statistics for the two models is 1.11 – 0 = 1.1. Since the p-value for 1.11 with 2 – 0 = 2 degrees of freedom is .57 > .05, we conclude there is no significant difference between the two models. This indicates that the interaction between all the variables does not make a significant contribution.
We next compare (CG, GT, CT) with (CG, GT). The difference in the statistics is 7.86 – 1.11 = 6.75, which yields a p-value of .034 on 4 – 2 = 2 degrees of freedom. This is a significant difference, and so we don’t consider (CG, GT) further.
We next compare (CG, GT, CT) with (CG, CT). The difference in statistics is 7.86 – 1.11 = 6.75, which yields a p-value of .106 on 4 – 2 = 2 degrees of freedom. This is not a significant difference, and so we now adopt the simpler model (CG, CT). This indicates that the interaction between G (gender) and T (therapy) does not make a significant contribution.
There is only one other qualifying model to consider, namely (T, CG). The difference in statistics is now 12.04 – 7.86 = 4.18, which yields a p-value of .0398 on 4 – 2 = 2 degrees of freedom. This is a significant difference, and so we reject the (T, CG) model. This indicates that the interaction between C (cure) and T (therapy) does make a significant contribution.
Conclusion
We conclude that (CG, CT) as the simplest (i.e. most parsimonious) model that significantly fits the data.
This indicates that there is an interaction between cure and gender as well as between cure and therapy. This can be seen by looking at the odds ratios of the observed data (see Figure 2).
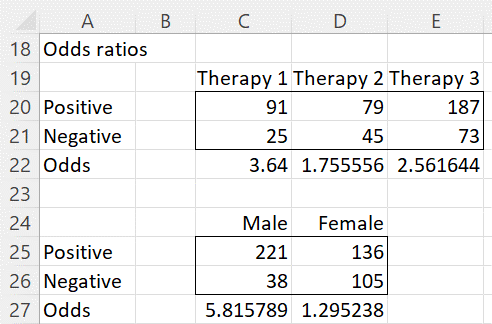
Figure 2 – Odds ratios
That there is an interaction between Cure and Therapy can be seen from the fact that the odds of a cure for therapy 1 is 91/25 = 3.64, while that for therapy 2 is 79/45 = 1.76. The odds ratio is 2.07, i.e. therapy 2 seems to be twice as effective as therapy 1.
That there is an interaction between Cure and Gender can be seen from the fact that the odds of a cure for males is 221/38 = 5.82, while that for females is 136/105 = 1.30. The odds ratio is 4.49, i.e. the therapies seem to be much more effective for men than for women.
Model coefficients
Finally, we calculate the coefficients of the log-linear model for (CG, CT)
using the following coding of the categorical variables:
tC = 1 if cured and = 0 otherwise
tG = 1 if male and = 0 otherwise
tT1 = 1 if therapy 1 and = 0 otherwise
tT2 = 1 if therapy 2 and = 0 otherwise
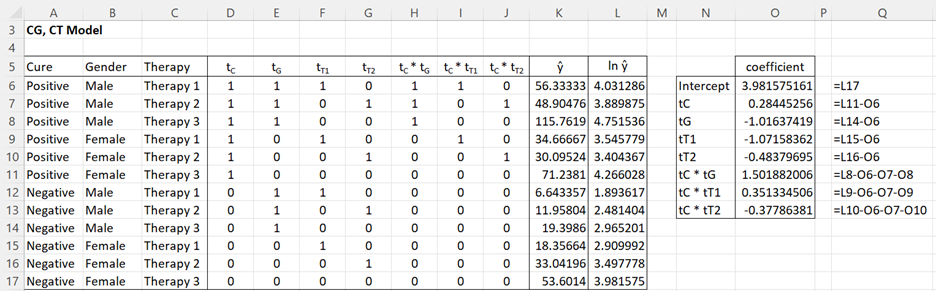
It is relatively easy to calculate the coefficients for this model, as described in Figure 3.
Figure 3 – Coefficients for (CG, CT) model
Alternatively, we can use Excel’s regression data analysis tool using L5:L17 as the Y range and D5:J17 as the X range. The coefficients outputted are the same as those given in Figure 3. The rest of the output from the data analysis tool should be ignored.
Thus the log-linear model is
![]()
Examples Workbook
Click here to download the Excel workbook with the examples described on this webpage.
Reference
Howell, D. C. (2010) Statistical methods for psychology (7th ed.). Wadsworth, Cengage Learning.
https://labs.la.utexas.edu/gilden/files/2016/05/Statistics-Text.pdf
what if there is a situation like, (CG, GT) and (CT, CT) are all not significantly different from (CG,GT,CT)? than what would be the model???
I meant, (CG, GT) and (CG, CT).. what if simpler form are both qualified to be in the model? Do we still use (CG,CT, GT)? or something else?
Hong,
I apologize for the delayed response.
If I understand correctly, then both (CG, GT) and (CG, CT) would be best-fit models since they would have the same number of factors.
Charles.
Dear Charles:
I have recently downloaded and installed your add-in for Excel, and I find it wonderful! Thank you.
Now, I am interested in exploring log-linear analyses for some data I have. BUT, although you explain here (in your webpage) how to perform the analysis… I can not find the proper module in Real Statistics.
I guess this is really simple, but I would like you to help me with this.
Thanks in advance:
Jose.
Hello Jose,
I am pleased that you like the Real Statistics software.
The approach for creating log-linear models is fully described on the website, but is not yet implemented in the Real Statistics software.
Charles
I am very grateful for your instructions on choosing models. I have followed the instructions very carefuly, but I must be confused with ‘We next compare (CG, GT, CT) with (CG, CT). The difference in statistics is 7.86 – 1.11 = 6.75, which yields a p-value of .106 on 4 – 2 = 2 degrees of freedom. This is not a significant difference, and so we now adopt the simpler model (CG, CT). This indicates that the interaction between G (gender) and T (therapy) does not make a significant contribution’. Is (CG, CT) not 5.592 according to the table?
Hello Dr Simon Sandett Namnyak,
Sorry for taking so long to respond. In this period I have been swamped with requests and so I haven’t been able to respond as rapidly as I would have liked.
The chi-square statistic for (CG, CT) is 5.592 as you have stated, but, perhaps I am missing something, I don’t see why you are asking the question.
When comparing the (CG, GT, CT) and (CG, CT) models there isn’t a significant difference, and so we eliminate the first of these models since it is more complex. Thus, in the context of (CG, GT, CT) and (CG, CT), we can drop the GT. This is all that is meant by my statement.
Charles